We are currently editing a special issue of Socius about the Challenge. For this special issue, we are striving for a standard of computational reproducibility, which means that other researchers should be about to recreate the results in all of the papers. Therefore, while the manuscripts have been undergoing peer review, we have also been attempting to replicate the results in each paper. This has turned out to be trickier than we expected. In this post, I’d like to briefly summarize what we’ve done so far, and then share a set of guidelines that we’ve developed and shared with our authors. If you have ideas for how these guidelines can be improved, please let us know. Ultimately, we hope that the guidelines will be a helpful resource for authors and editors who wish to promote computational reproducibility, either in their own work or the work of others.
Our replication efforts have been spearheaded by David Liu, and this work will be part of his senior thesis in Princeton’s Department of Computer Science. In attempting to replicate the results of each paper, David has noticed helpful things that some authors have done, and he’s found some problems that come up over and over. Therefore, when we sent back decisions on the manuscripts, we also sent the feedback below on code. Just as authors have to revise and resubmit their manuscripts, for the special issue, authors will have to revise and submit their code. These guidelines are intended to help with that process.
Background behind reproducibility guidelines
First, we’d like to step back from the details to describe the high-level goal. We want your articles to be computationally reproducible, which means that another researcher could regenerate the results in your paper using the Challenge data, your code, and any additional data that you have created. Computational reproducibility will increase the impact of your work individually, and it will increase the contribution of the Challenge collectively.
As we’ve learned during this first round of reviews, the goal of computational reproducibility is widely shared by scientists, easy to state, and tricky to achieve. Based on what we’ve learned from your code, our thinking on how to achieve this goal issues has evolved. In particular, we’ve been very influenced by the idea of a “research pipeline” described by Peng and Eckel (2014), which is nicely captured by this figure: http://bit.ly/2qrTWXK.
The goal of this document is to provide you with guidelines that support computational reproducibility of your entire research pipeline, which goes from raw data to final output. You don’t have to follow these guidelines exactly; if you devise a system that you think is better, you are welcome to use it. But, if you have no system in place, we are going to strongly encourage that you adopt these guidelines.
The Guidelines
The most important thing to keep in mind is that we are asking you to create one single script named “run_all” that executes all the necessary files to go from the raw data to the final results. One way to do this is to write a bash script that calls the submission files in sequence. An example of a simple bash script is shown below:
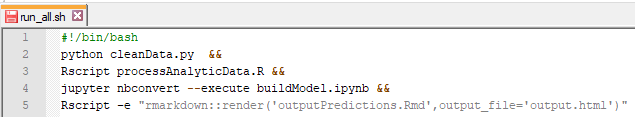
Running the above script will execute each line, one after another. Note that the screen shot includes examples for many common languages. More background information on writing bash scripts is available at: https://ryanstutorials.net/bash-scripting-tutorial/bash-script.php. Of course, you may write the run_all script in the language of your choice so long as it can be executed from the command line.
While you are creating this script, we think it will be helpful to organize your input files, intermediate files, and output files into a standard directory (i.e., folder) structure. We think that this structure would help you create a modular research pipeline; see Peng and Eckel (2014) for more on the modularity of a research pipeline. This modular pipeline will make it easier for us to ensure computational reproducibility, and it will make it easy for other researchers to understand, re-use, and improve your code.
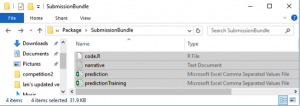
Here’s a basic structure that we think might work well for this project and others:
data/
code/
output/
README
LICENSE
In the data/ directory you can include:
- background.csv (this should not actually be included because of privacy constraints, but we will put it here)
- train.csv (this should not actually be included because of privacy constraints, but we will put it here)
- Supplemental materials such as metadata files, the constructed-data dictionary, the machine-readable codebook.
- Data that you have collected or created, such as a csv file that you manually created that has your MSE scores on the holdout data and/or an analytic dataset created by your code.
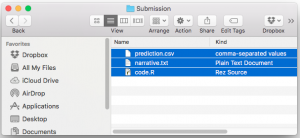
In your code/ directory you can include:
- Executable “run all” script that when run goes from raw inputs all the way to final outputs (for this script we encourage you to think about the research pipeline idea from Peng and Eckel 2014: http://bit.ly/2qrTWXK)
- Source code files each with a useful header (see FAQ).
- Package requirements
In your output/ directory you can include:
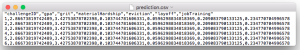
- prediction.csv
- A subdirectory for tables
- A subdirectory for figures (we also recommend including all data files that can be used to recreate the figures; see rule 7 of Sandve et al. 2013)
In addition to these three main directories, you should also include a README file and LICENSE file. We have more information about these files in the FAQ below. We hope that these guidelines are help, and please let us know if you have any questions.
Code Resubmission Process
Once you think you are ready to resubmit, here’s a checklist that you can follow to help ensure that your work will be computationally reproducible:
- I have written the kind of README file that I would like to read (see FAQ below)
- Each code file that I’ve written has a header that will be helpful (see FAQ below)
- I’ve run the submission and I can get from raw files to final output using only materials in my directories. Then, I’ve done this again and I get the same result. This second step helps check for problems with seeding.
- I’ve considered refactoring my code (see FAQ below)
Finally, when you resubmit, we ask that you include a revision memo about the code, just as you will about the manuscript. This revision memo should summarize changes that you have made. In this revision memo, please also include a rough estimate of the cumulative amount of time it took you to comply with these guidelines. We are asking for this time estimate because one objection to computational reproducibility is that it is too burdensome for authors and we would like to assess this empirically. Finally, please include any suggestions for how this process could have been easier or more efficient.
F.A.Q.
What should go in the README file?
The README file should provide an overview of your code. For example, it could include a diagram showing the different pieces of their code, their inputs and their outputs. If relevant, please include expected warnings when executing the code. Mention any provided “intermediate results” readers can utilize to decompose the submission into smaller pieces.
The README should also include something about your computing environment and expected run time; general terms are appropriate here. For example: “I ran this on a modern laptop (circa 2016) and it ran in a few minutes.” or “This code ran on high-performance cluster and took one week.” Finally, please clearly cite any open sourced content utilized in the submission, such as resources shared in the FFC blog or more general packages distributed in the computation community.
What headers should be included at the top of each piece of code?
Based on the ideas in Nagler (1995), we think the following elements should be included at the top of each piece of code:
- Purpose (in 140 characters or less)
- Inputs
- Outputs
- Machine used (e.g., laptop, desktop, cluster)
- Expected runtime (e.g., seconds, minutes, hours, days, etc)
- Set the seed at the beginning of each file (see rule 6 of Sandve et al. 2013)
- All the package include statements (e.g., “library(ggplot2)” in R)
If you would like to deviate from this standard, please contact us.
How can I make my code easier to read?
It is hard to offer general advice, but one thing that we can recommend is at the end of the process take some time to refactor your code (https://en.wikipedia.org/wiki/Code_refactoring). In our experience, code evolves over the course of a project, and at the end it can be helpful to refactor in order to clean up the structure, improve variable names, and promote modularity.
Even if you don’t refactor your code, please include additional comments to helper functions and code segments that may be obscure to new readers.
What is our standard for computational reproducibility for the special issue?
Our standard for computational reproducibility for this special issue is that we should be able to take whatever code and data you submit, add the Fragile Families Challenge data file, and then reproduce all of the figures in your paper, all of the tables in your paper, and your predictions.csv file.
What is not included in our standard for computational reproducibility for the special issue?
We will not attempt to completely recreate your analysis from the written materials. Also, we will not verify that your description in the paper matches the code. For example, if the paper says that you use logistic regression to generate your predictions, we will not verify that the code also uses logistic regression. Further, we will not verify the information that you have provided from external sources. For example, if you write in the paper that your submission was 10th on the leaderboard, we will not verify this fact. Finally, we will not verify any of the numbers that are included in the text of the manuscript. For example, we would not verify a claim in the text such as: dropping variables with no variation removes 10% of variables. As we hope this list illustrates, our standard of computational reproducibility is in fact quite limited.
What license should I use?
We strongly recommend the MIT license. You can find it here: https://opensource.org/licenses/MIT. Simply replace with 2018 and with the name of all co-authors of the paper, in the order they are listed in the paper. If you would like to use some other license, please contact us.
What should I read to learn more about computational reproducibility?
Here’s a partial list. If we’ve left off a good resource, please let us know (fragilefamilieschallenge@gmail.com).
Nagler (1995) “Coding Style Good Computing Practices” PS: Political Science & Politics. (open access version)
Peng and Eckel (2009) “Distributed Reproducible Research Using Cached Computations” Computing in Science & Engineering.
Sandvae et al (2013) “Ten Simple Rules for Reproducible Computational Research” PLOS Computational Biology.
Stodden et al (2016) “Enhancing reproducibility for computational methods” Science.